Full Research Projects
-
Project #1 (Target Identification) - MYC Signaling in Poor Prognosis Luminal B Breast Cancer
Lead PI: Ernest Martinez, Ph.D.
Co-Lead PI: Veronica Jones, M.D.Abstract
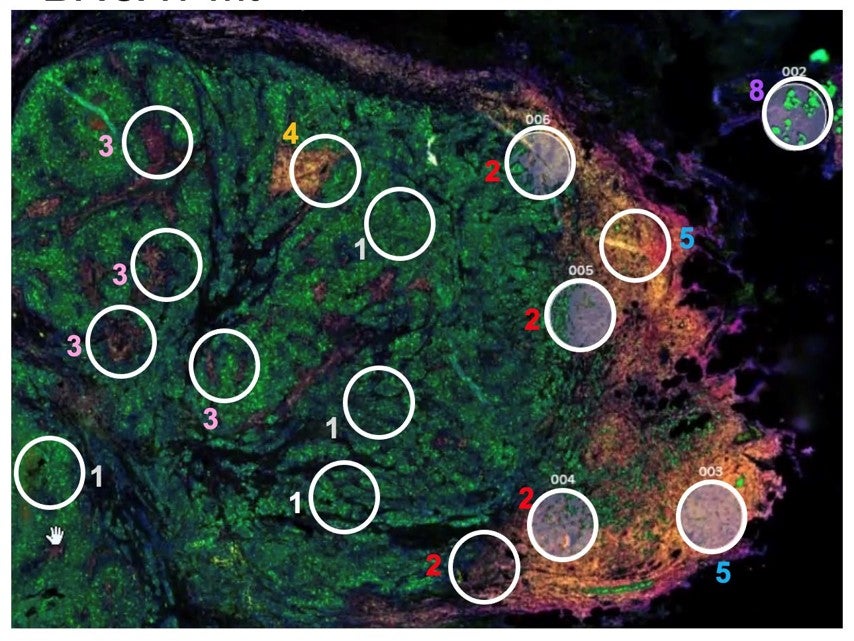
Most luminal B breast cancers (LBBC; ER+, HER2-wt, Ki67>14%) carry a good prognosis. However, approximately 20% of LBBC are highly aggressive and resistant to current therapies. There have been many failed attempts to therapeutically target MYC. These attempts have failed, in part, due to our current inability to separate the cancer-promoting activities of MYC, mechanistically or therapeutically, from its normal essential cellular functions. In preliminary data, we show that the cancer-transforming ability of MYC is dependent on three lysine (K) residues of MYC (K149, K158, and K323) that are major substrates for acetylation by the histone acetyltransferases (HATs) p300 and GCN5. Guided by our preliminary data, here we aim to dissect the cofactors and molecular mechanisms by which these MYC acetyl-K (AcK) residues promote cell transformation and initiation and progression of LBBC. Our long-term goal is to identify new “druggable” targets to improve survival of women most affected by aggressive LBBC. Guided by our preliminary data, we hypothesize that a gene-selective MYC-AcK signaling pathway drives the aggressive tumor cell biology of therapy-resistant LBBC and involves transcription cofactors and epigenetic coregulators that “write” and/or “read” AcK marks on MYC and histones, perhaps including cofactors co-overexpressed with MYC in LBBC, such as PIN1, GCN5, p300, and/or YEATS2. Here, we will investigate the role of MYC-AcK dependent signaling in luminal mammary epithelial cell transformation and aggressive biology of LBBC. We will leverage Dr. Jones's fully annotated tissue collection of 349 women who were treated for Stage 2/3 LBBC. Aim 1 will define the impact of MYC-AcK dependent signaling in mammary epithelial cell transformation and in the aggressive biology of LBBC. Aim 2 will characterize the molecular mechanisms of MYC-AcK dependent gene regulation in transformed mammary epithelial cells and in LBBC from patients.
-
Project #2 (Drug Development) - Development of Inhibitors of the PIN1 Oncoprotein in Pancreatic Cancer
Lead PI: Maurizio Pellecchia, Ph.D.
Co-Lead: Mustafa Raoof, M.D., M.S.
Co-Investigators: Gregor Blaha, Ph.D., David Horne, Ph.D.Abstract
Pancreatic cancer is highly aggressive and has an extremely low 5-year survival rate. Pancreatic ductal adenocarcinoma (PDAC) is notoriously resistant to the majority of treatments including cytotoxic chemotherapy, targeted agents, and immuno-therapy. Treatment resistance has been linked to tumor heterogeneity, limited tissue penetration of drugs, and an immunosuppressive tumor microenvironment (TME). PIN1 is a cis-trans prolyl isomerase that controls proline-mediated phosphorylation signaling events that is overexpressed both in pancreatic cancer cells and cancer-associated fibroblasts. PIN1 overexpression is a major contributor to tumorigenesis, activating several oncoproteins, including proteins in the KRAS pathway, 17 and simultaneously inactivating several tumor suppressors. PIN1 promotes an immunosuppressive/treatment-resistant TME, by up-regulating PD-L1 (programmed cell-death receptor-1) Guided by our resources and preliminary data, we propose a collaboration between UCR and CoHCCC to optimize and develop a potent and selective PIN1 inhibitor for treatment of pancreatic cancer. Aim 1 will design, synthetize, and iteratively optimize novel, drug-like PIN1 targeting agents. Here we propose to optimize Dr. Pellecchia's initial discoveries, iteratively refining the structures of our hit compounds for potency and efficacy, with particular attention to drug-like properties of the compounds (Dr. Pellecchia, UCR). The hit-to-lead optimization phase will be fully integrated with cellular pharmacology (Dr. Pellecchia, UCR) studies, and in vitro ADME properties measurements (Dr. Horne, CoH), that will be iteratively assessed. In addition, the CoH state-of-the-art core facilities will be deployed to profile the most active agents against multiple cell lines, and to conduct high throughput cell efficacy studies in combinations. Aim 2 will study the mechanism of action and efficacy of most promising agents in cellular and animal models of pancreatic cancer. We will assess the pharmacokinetics properties of refined agents in mice and test their efficacy in animal models of pancreatic cancer.
-
Project #3 - Capacity Building Clinical Trial of Metformin Against Inflammation in Insulin-Resistant Breast Cancer Survivors
Lead PI: Kendrick Davis, Ph.D.
Co-Lead: Victoria Seewaldt, M.D.Abstract
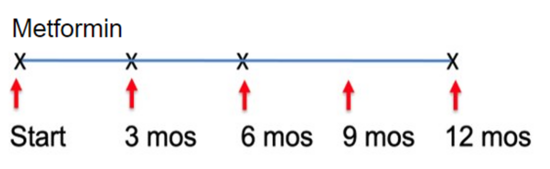
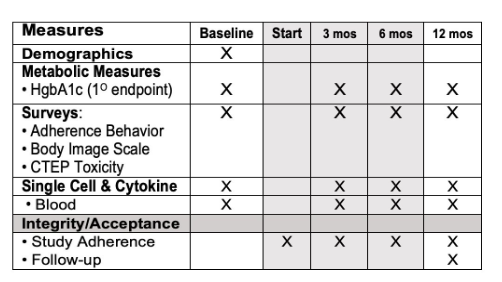
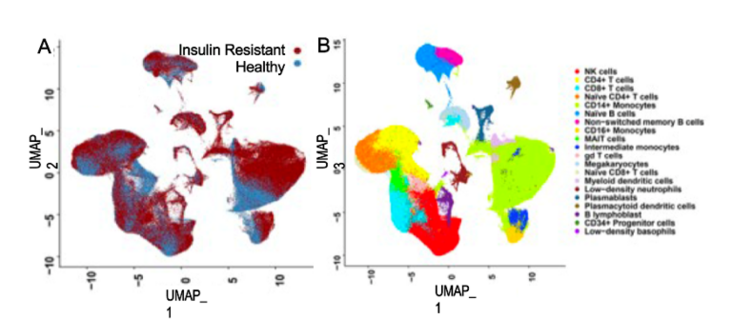
This study aims to implement a readily available, inexpensive, and safe measures to restore metabolic health and reverse accelerated aging in young adults (aged 18-35) in the rural farming Coachella Valley in Southern California. Worldwide, 34% of young adults are metabolically unhealthy, and poor metabolic health is associated with accelerated aging, decreased quality of life, and enormous economic burden for the healthcare system. Our studies show that insulin resistance leads to epigenetic damage that drives inflammation and accelerated aging. Without intervention, unhealthy young adults face a lifetime of chronic disease and early death. Effective, affordable, and scalable intervention strategies are needed to reverse metabolic disease and aging in young adults. Our proposed study will investigate the impact of metformin to reverse accelerated aging. We chose metformin because it is 1) are readily available and affordable, 2) known to restore metabolic health, and 3) shows promise to slow aging. Alone, metformin is inexpensive and safe (even in pregnancy); and it slows aging (slows telomere shortening, inflammation, DNA damage). Metformin also 1) decreases hunger by increasing sensitivity to leptin and production of GLP-1, and 2) increases delivery of glucose to muscle. Guided by our preliminary studies and supported by a concept-mapping implementation strategy, we will test in young insulin-resistant adults (aged 18-35) the hypothesis that 12-month metformin will restore metabolic health and reverse accelerated aging. We will test mechanistic markers of aging. Aim 1 will test whether metformin reverse pre-diabetes and inflammation. Aim 2 will test whether restoration of metabolic health (HgbA1c<5.7) reverse epigenetic damage and accelerated aging. Evaluate 0, 12 months. Aim 3 will test whether restoration of metabolic health (HgbA1c<5.7) improves mood and wellness using conventional measures and an Emoji evaluation tool.

Pilot Research Projects
-
Pilot #2 - Control of Protein Synthesis by eIF4A1 and mRNA m6A Modification in Acute Myeloid Leukemia
Lead PI: Seán O’Leary, Ph.D.
Co-Lead: Rui Su, Ph.D.Abstract
Cancer cells require sustained high levels of protein synthesis for oncogenesis and disease progression. To achieve this, the cellular protein synthesis (“translation”) machinery is dysregulated to facilitate the biochemical and physiological demands of the cancer cell. Acute myeloid leukemia (AML) is a common and fatal hematopoietic malignancy. Despite improved therapeutic options, over 70% of AML patients cannot survive beyond 5 years. This underscores a critical need for more effective approaches to treat AML. Emerging evidence indicates that aberrant translation is a hallmark of leukemia and targeting mRNA translation represents a promising strategy to combat AML. Translation initiation factor 4F (eIF4F) is a heterotrimeric protein complex, including a cap-binding subunit eIF4E, an RNA-binding/scaffolding subunit eIF4G, and a DEAD-box RNA helicase eIF4A7. eIF4F recognizes the mRNA 5ʹ cap, enabling mRNA recruitment to the ribosome. This complex has been recognized as an important driver of oncogenesis; translational deregulation significantly increases its activity in cancer, including leukemia. eIF4F is thus a major cancer chemotherapeutic target. Inhibitors of all its subunits have offered significant promise as anticancer agents. N6-Methyladenosine (m6A), the most prevalent modification in mRNAs, plays a fundamental role in regulating mRNA translation, and its dysregulation might lead to oncogenesis. In the best-characterized mechanism. . Here, we propose an integrated in-vivo and in-vitro study that comprehensively quantifies the impact of mRNA m6A modification on eIF4A1/eIF4F function, from single-molecule level, through in-vitro biochemistry, to the realm of cellular and animal physiology. Aim 1 we will define the role of eIF4A1 in AML pathogenesis. Aim 2 will dissect the biochemical impact(s) of mRNA m6A and its “reader” and “writer proteins IGF2BP1 and METTL14 on eIF4A1/eIF4F function. Our ultimate aim is to develop less toxic and more effective treatments for AML.
-
Pilot #3 - Utilizing RELM-alpha-/-(M2 Macrophage-Polarized) Mice to Develop Novel Desmoplastic Models and Targeted Therapies for Pancreatic Cancer
Lead PI: Meera G. Nair, Ph.D.
Co-Lead: Edwin R. Manuel, Ph.D.Abstract
Pancreatic ductal adenocarcinoma (PDAC) is projected to become the second-leading cause of cancer-related death by 2030. Desmoplasia, or fibrosis, is a hallmark of the PDAC tumor microenvironment (TME) and is comprised of a dense extracellular matrix (ECM) containing copious amounts of hyaluronic acid and collagens. Desmoplasia negatively affects prognosis because it: 1) acts as a biophysical barrier to therapy, 2) increases interstitial fluidic pressure leading to blood vessel compression and 3) initiates signal cascades that suppress immunity and promote invasiveness. Biochemical cues initiated by the TME ultimately contribute to proliferation, migration, invasion, angiogenesis and apoptotic resistance. Effective methods to overcome desmoplasia in PDAC, in order to improve drug delivery and efficacy, continues to be a significant unmet need. Development of novel models that physiologically recapitulate fibrosis in human PDAC would accelerate evaluation of ECM-targeting strategies. There is evidence that poor prognosis PDAC is predicted by a greater prevalence of tumor-associated macrophages (TAMs) that are primarily M2-polarized (pro-tumor) TAMs are one of the most abundant immune subsets in the PDAC stroma and promote fibrosis, tumorigenesis, immune escape, metastasis and therapeutic resistance. Strategies to eliminate or reprogram TAMs into a more M1 (anti-tumor) phenotype is currently an area of intense research. In this application, we propose complementary studies that will leverage the expertise of Drs. Meera G. Nair (UCR) and Edwin R. Manuel (COH) to develop novel tools and therapies that will aid in minimizing desmoplasia and M2 TAMs in PDAC. Aim 1 will characterize PDAC tumors implanted in M2 macrophage-driven RELM-deficient mice. Aim 2 will target M2 TAMs using a bacterial-based, shRNA plasmid delivery system.